Introduction to Bioinformatics
📘 This content is part of version: v1.0.0 (Major release)What is bioinformatics?¶
The field of bioinformatics deals with research, development and application of computational tools and approaches for expanding the use of molecular, biological, medical, behavorial or health data. It is also concerned with means to acquire, store, organize, archive, analyze or visualize such data. Bioinformatics is an interdisciplinary field at the intersection of, among others, biology, computer science and statistics.
The first activities in bioinformatics trace back to the 1950s, when researchers such as Frederick Sanger started to experimentally elucidate the sequences of proteins and - later - DNA. Margaret Dayhoff was among the first to develop algorithms to analyse such sequences Dayhoff & Ledley, 1962. The term “bioinformatics” was subsequently coined by Paulien Hogeweg and Ben Hesper in 1970 Hesper & Hogeweg, 2021, to describe “the study of informatic processes in biotic systems”, and gradually became used to denote the entire field.
Research in bioinformatics really took off in the 1990s, with the introduction and wide-spread use of -omics technologies (DNA sequencing, gene expression microarrays, mass spectrometry for proteins and metabolites, etc.) and the Human Genome Project. Much of the resulting data, as well as the knowledge gained in these experiments, was made freely available for researchers in the form of computer databases. The processing and analysis of these large volumes of data on molecules and their interactions required dedicated algorithms and sofware to be developed. Bioinformatics thus further developed into a thriving research area in its own right, providing the computational methods that are by now essential for most biological research. Computational biologists these days even address complex biological and evolutionary questions without going into the lab, by integrating and interrogating existing data in novel ways.
Teaching bioinformatics¶
Bioinformatics can be taught at a wide range of levels, from high school biology students to BSc students in biological or biotechnological programmes and even MSc or PhD students in bioinformatics. While researchers in bioinformatics or computational biology will need to develop a sound basis in statistics, programming and data analysis/machine learning, a far broader range of life scientists - the target audience of this book - will make use of bioinformatics methods as part of their everyday work or research, and mostly require practical knowledge. For these students, detailed understanding of the underlying mathematics and computer science is less relevant; they should however realise what main methods and databases are available, what their specific uses and limitations are, and how they should interpret the outcomes of computational analyses.
To support life science students to reach these learning goals, we developed a BSc course “Introduction to Bioinformatics” at Wageningen University. We structured it around the “journey” a researcher would take to study an unknown sequence, as illustrated in Figure Figure 1.
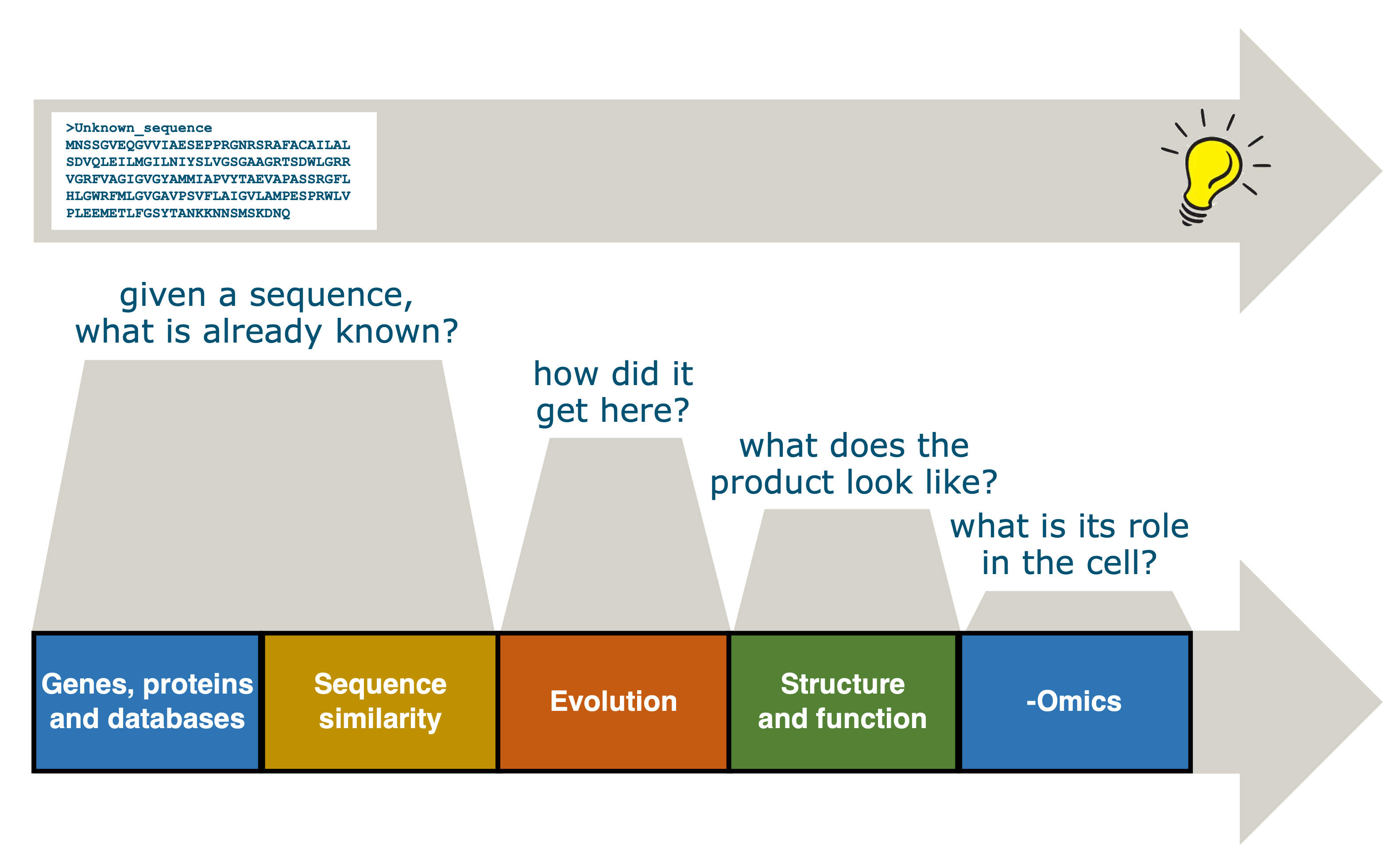
Figure 1:Overview of the chapters in this book. Credits: CC BY 4.0 Ridder et al., 2024.
Why a new book?¶
Bioinformatics is a fast moving field with a constant stream of new techniques, tools and relevant biological insights. While a number of undergraduate text books on the topic are available, they generally struggle to keep up with this high rate of change. As teachers of a BSc-level introductory course on bioinformatics, we struggled each year to collect a relevant set of reading material on the topics we intend to cover. Where one book might fit one week, the chapters relevant to another week might have been outdated, forcing us to use different books. In addition, the level of detail in the books did not always correspond with the level of understanding we aim to achieve in students.
We therefore decided to bundle current knowledge and our expertise in the form of a book, tailored to the level that we aim to achieve in our BSc course. The five steps along the journey above are reflected in a setup with five chapters, that each contain basic text, various boxes with different types of material (see below), practical exercises and references:
- Genes, Proteins, Databases, Genome annotation
- Alignment, Sequence search, Primer design
- Pylogenetics and tree building
- Protein structure prediction
- Omics data analysis
After some years of development and streamlining, we have now reached the stage that we feel it is ready to release it to the community as an online textbook, free for use. We hope you find it useful!
Be aware that the practical exercises mostly rely on online databases and tools, that can be inacessible at times and regularly change their layout, operation or use policies; each fall, we revisit these exercises to ensure they work for our class, but at other times you may run into issues. Also note that image enlargement is currently not built into the reader. To enlarge images, please right-click an image and select “Open image in new tab” (browser dependent).
As with any textbook, you might find errors or omissions.
If you do find any, please contact us at
introduction@bioinformatics
Dick de Ridder, Anne Kupczok, Rens Holmer, Freek Bakker, Justin van der Hooft, Judith Risse, Jorge Navarro and Tomer Sardjoe (Wageningen, September 2025)
Reading guide¶
The text is decorated with different types of boxes that show additional information or are used to highlight and clarify important aspects. Yellow and blue boxes are part of the exam material for this course, green boxes provide useful additional information or links.
References¶
- Dayhoff, M. O., & Ledley, R. S. (1962). Comprotein: a computer program to aid primary protein structure determination. AFIPS ’62, Proceedings of the December 4-6, 1962 Fall Joint Computer Conference, 262–274. 10.1145/1461518.1461546
- Hesper, B., & Hogeweg, P. (2021). Bio-informatics: a working concept. A translation of “Bio-informatica: een werkconcept.” 10.48550/arXiv.2111.11832
- Ridder, D. de, Kupczok, A., Holmer, R., Bakker, F., Hooft, J. van der, Risse, J., Navarro, J., & Sardjoe, T. (2024). Self-created figure.